What is Blast2KEGG
|
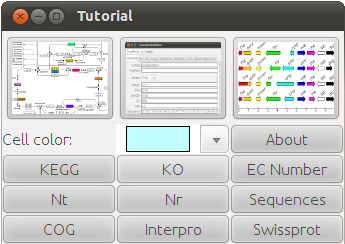
Blast2KEGG starts with a tutorial window. In this window, you need to choose files to upload and set the color you want to paint on KEGG viewer. After all files are uploaded, you can go on checking genes mapped to pathway use the big button on the left, continue editing the annotation using the big button in the middle, or browser genes using big button on the right (see figure below). |
||||||||||||||||||||||
|
||||||||||||||||||||||
|
Blast2KEGG Command-line Specification It is much more convenient to input sequences and annotations into Blast2KEGG from command line, all files at one time. The following options are currently available:
|
||||||||||||||||||||||
|
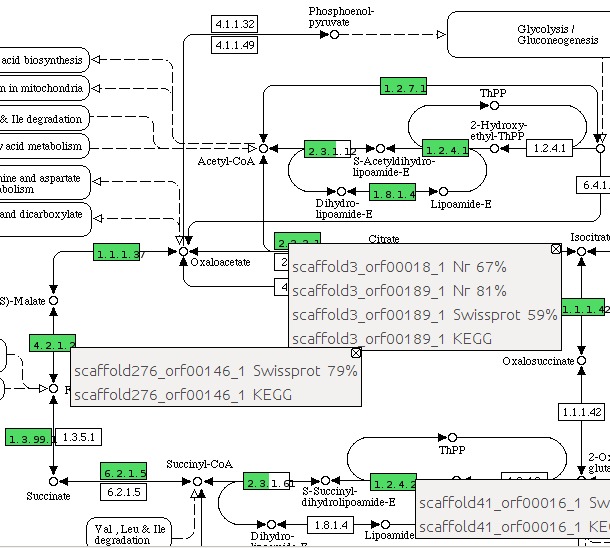
The KEGG viewer contains the pathway maps downloaded from http://www.genome.jp/kegg/. You can use the drop list to choose a map. The EC numbers existed in the uploaded annotations are highlighted according to their similarities. When mouse moves over a highlighted EC number, a tooltip pops up to show the coresponding genes and the matches to the databases. This tooltip can be fixed by clicking the right mouse button (see figure below). |
||||||||||||||||||||||
|
||||||||||||||||||||||
|
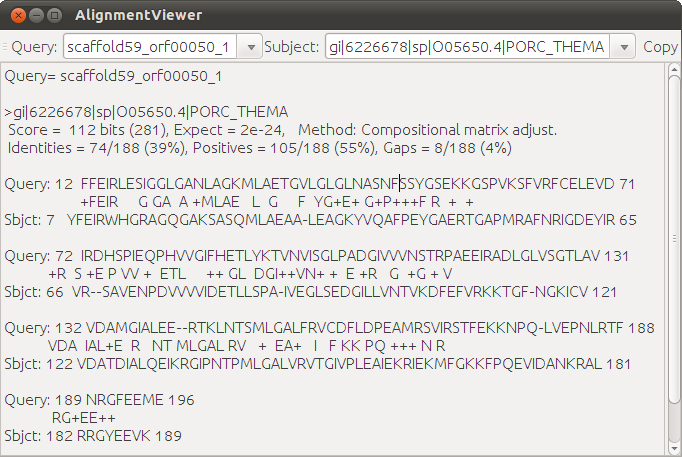
On left click the highlighted EC number, a dialog pops up to show the alignments between the query genes and the databases (see figure below). |
||||||||||||||||||||||
|
||||||||||||||||||||||
|
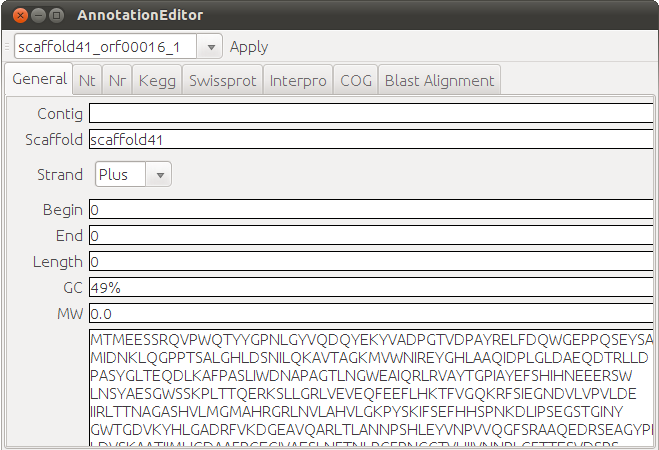
An annotation editor can be accessed from the tutorial window, or the Edit button in KEGG viewer. It keeps a record of the annotations loaded from various databases. User can also input their own annotaions manually, and the annotations can be saved for future use (see figure below). |
||||||||||||||||||||||
|
||||||||||||||||||||||
|
The Genome Browers is still under development, it will act as a fast approach to connect pathways and gene clusters in the future. |
||||||||||||||||||||||
|